 A primary goal in the field of neuroscience is understanding how the brain controls movement. By improving pose estimation, neurobiologists can more precisely…
A primary goal in the field of neuroscience is understanding how the brain controls movement. By improving pose estimation, neurobiologists can more precisely…
A primary goal in the field of neuroscience is understanding how the brain controls movement. By improving pose estimation, neurobiologists can more precisely quantify natural movement and in turn, better understand the neural activity that drives it. This enhances scientists’ ability to characterize animal intelligence, social interaction, and health.
Columbia University researchers recently developed a video-centric deep learning package that tracks animal movement more robustly from video, which helps:
- obtain reliable pose predictions in the face of occlusions and dataset shifts.
- train on images and videos simultaneously, while significantly shortening training time.
- simplify the software engineering needed to train models, form predictions, and visualize the results
Named Lightning Pose, the tool trains deep learning models in PyTorch Lightning on both labeled images and unlabeled videos, which are decoded and processed on the GPU using NVIDIA DALI.
In this blog post, you’ll see how contemporary computer vision architectures benefit from open-source, GPU-accelerated video processing.
Deep learning algorithms for automatic pose tracking in video have recently garnered much attention in neuroscience. The standard approach involves training a convolutional network in a fully supervised approach on a set of annotated images.
Most convolutional architectures are built for handling single images and don’t use the useful temporal information hidden in videos. By tracking each keypoint individually, these networks may generate nonsensical poses or ones that are inconsistent across multiple cameras. Despite its wide adoption and success, the prevailing approach tends to overfit the training set and struggles to generalize to unseen animals or laboratories.
An efficient approach to animal pose tracking
The Lightning Pose package, represented in Figure 1, is a set of deep learning models for animal pose tracking, implemented in PyTorch Lightning. It takes a video-centric and semi-supervised approach to training of the pose estimation models. In addition to training on a set of labeled frames, it trains on many unlabeled video clips and penalizes itself when its sequences of pose predictions are incoherent (that is, violate basic spatiotemporal constraints). The unlabeled videos are decoded and processed on the fly directly on a GPU using DALI.
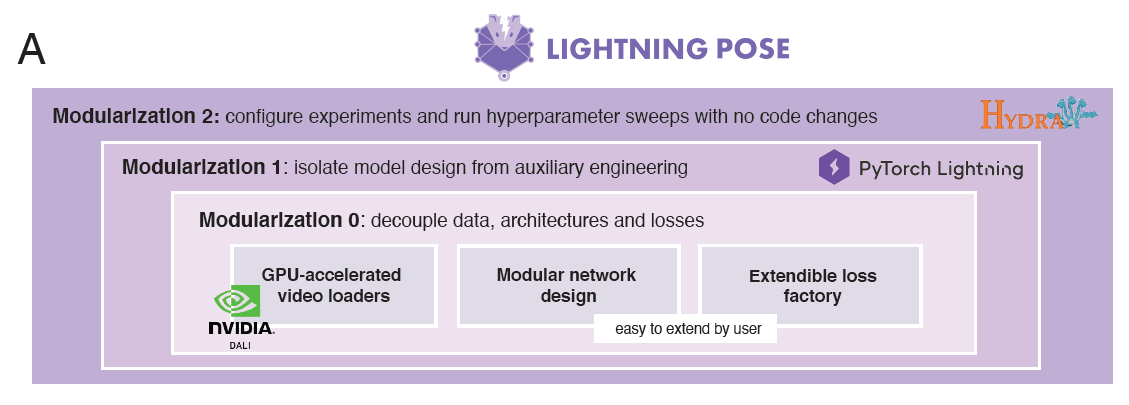
During training, videos are randomly modified, or augmented, in various ways by DALI. This exposes the network to a wider range of training examples and prepares it better for unexpected systematic variations in the data it may encounter when deployed.
Its semi-supervised architecture, shown in Figure 2, learns from both labeled and unlabeled frames.
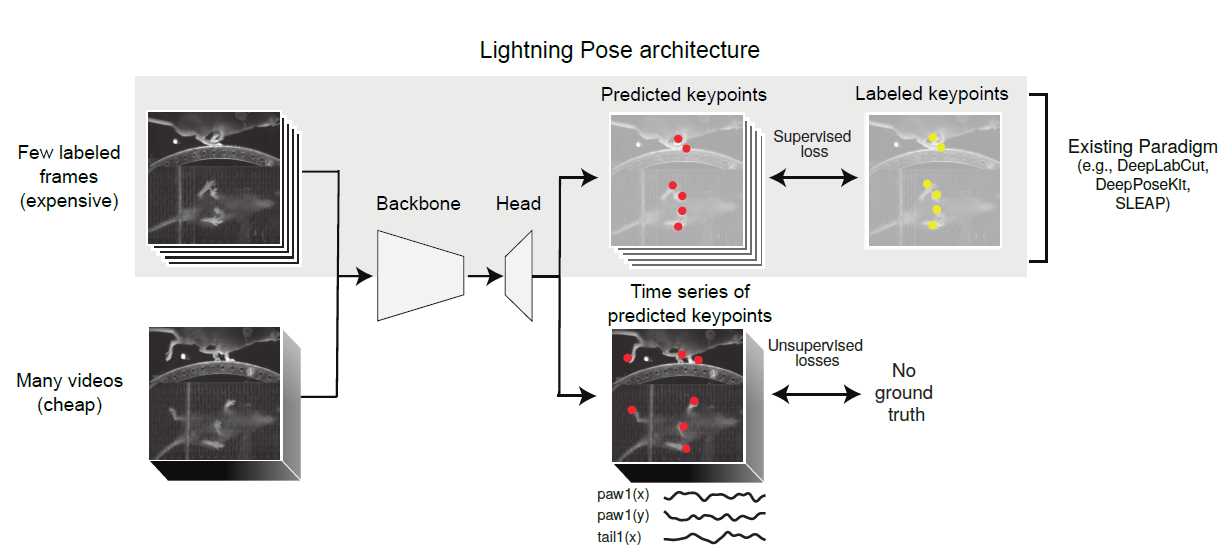
Lightning Pose results in more accurate and precise tracking compared to standard supervised networks, across different species (mice, fish, and so on) and tasks (full-body locomotion, eye tracking, and so on). The traditional fully supervised approach requires extensive image labeling and struggles to generalize to new videos. It often produces noisy outputs that hinder downstream analyses.
Its new pose estimation networks generalize better to unseen videos and provide smoother and more reliable pose trajectories. The tool also enhances robustness and usability. Through semi-supervised learning, Bayesian ensembling, and cloud-native open-source tools, models have lower pixel errors compared to DeepLabCut (with as few as 75 labeled frames). Lightning Pose estimation improves by 40, lowering pixel error and average keypoint pixel error across frames (DeepLabCut 14.60±4).
The clearest gains were seen in a mouse pupil tracking dataset from the International Brain Lab, where, even with over 3,000 labeled frames, the predictions were more accurate, and led to more reliable scientific analyses.
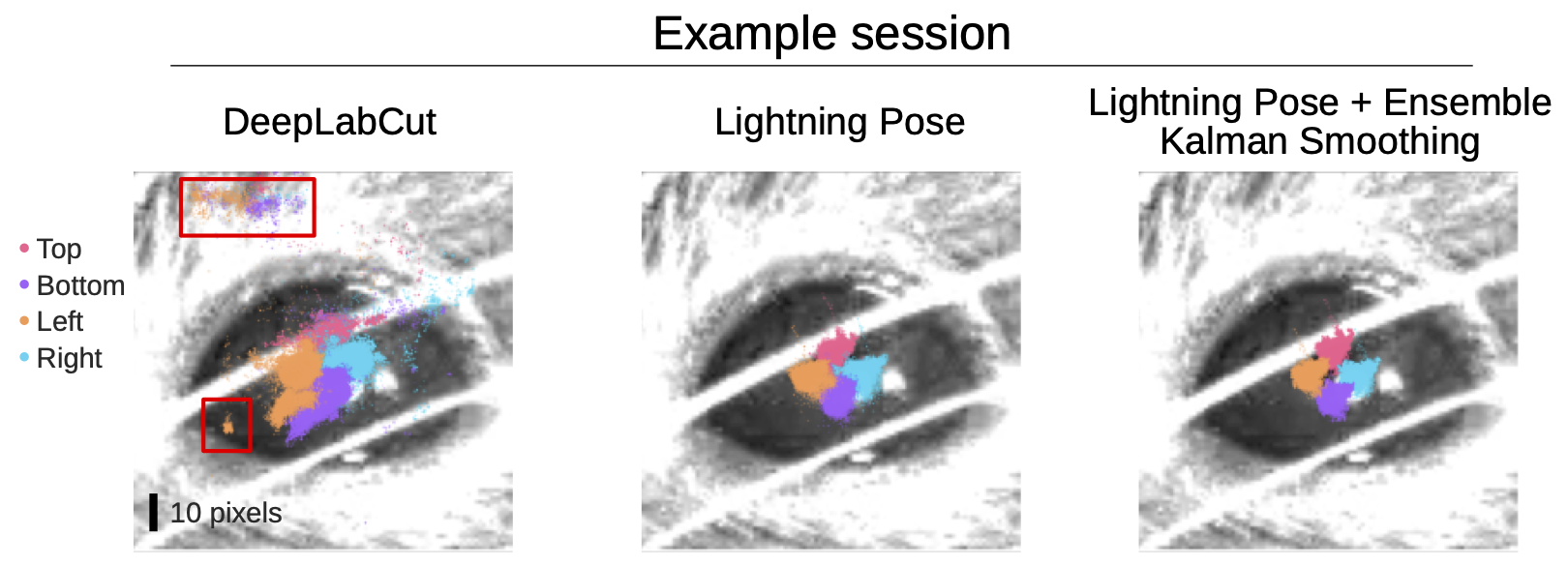
Figure 3 shows the tracking top, bottom, left, and right corners of a mouse’s pupil during a neuroscience experiment. On the left, the DeepLabCut model provides a significant number of predictions in implausible parts of the image (red boxes).
The center shows Lightning Pose predictions and the right, combines Lightning Pose with the authors’ Ensemble Kalman smoothing approach. Both Lightning Pose approaches nicely track the four points and predict them in plausible areas.
Improved pupil tracking in turn exposes stronger correlations with neural activity. The authors performed a regression between neural activity and tracked pupil diameter across 66 neuroscience experiments, and found that the model outputs were decoded more reliably from brain activity.
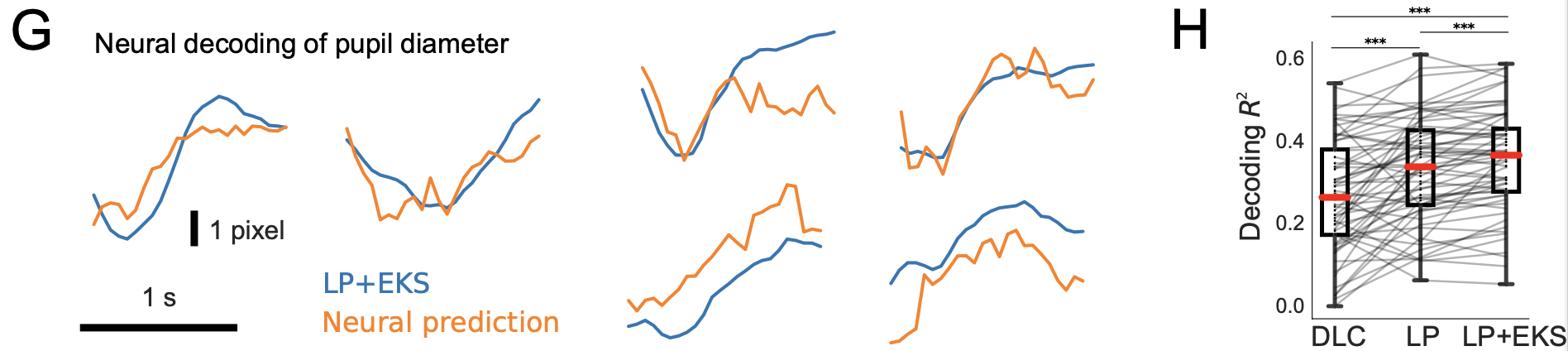
Figure 4 shows pupil diameter decoding from brain recordings. The left side of Figure 4 graphs pupil diameter time series derived from a Lightning Pose model (LP+EKS; blue), and the predictions from applying linear regression to neural data (orange).
The right side of Figure 4 shows R2 goodness-of-fit values quantifying how well pupil diameter can be decoded from neural activity. As shown, Lightning Pose and the ensemble version produce significantly better results DLC R2=0.27±0.02; LP 0.33±0.02; LP+EKS 0.35±0.02.
The following video shows the robustness of the predictions for a mouse running on a treadmill.
Improving the image-centric approach to convolutional architectures with DALI
Applying convolutional networks to videos presents a unique challenge: these networks typically operate on individual images. Despite the growing computational power of deep learning accelerators, such as new GPU generations, Tensor Cores, and CUDAGraphs, this image-centric approach has remained largely unchanged. Current architectures require videos to be split into individual frames during pre-processing, where they are often saved on a Disc for later loading. These frames are then augmented and transformed on the CPU before being fed to the network waiting on the GPU.
Lightning Pose leverages DALI for GPU-accelerated decoding and processing of videos. This stands in contrast to most computer vision deep learning architectures, such as ResNets and Transformers, that typically operate only on single images. When applied sequentially to videos, these architectures (and the popular neuroscience tools of DeepLabCut and SLEAP that are based on them) often form discontinuous predictions that violate the laws of physics. For example, an object jumping from one corner of a room to another, in two consecutive video frames.
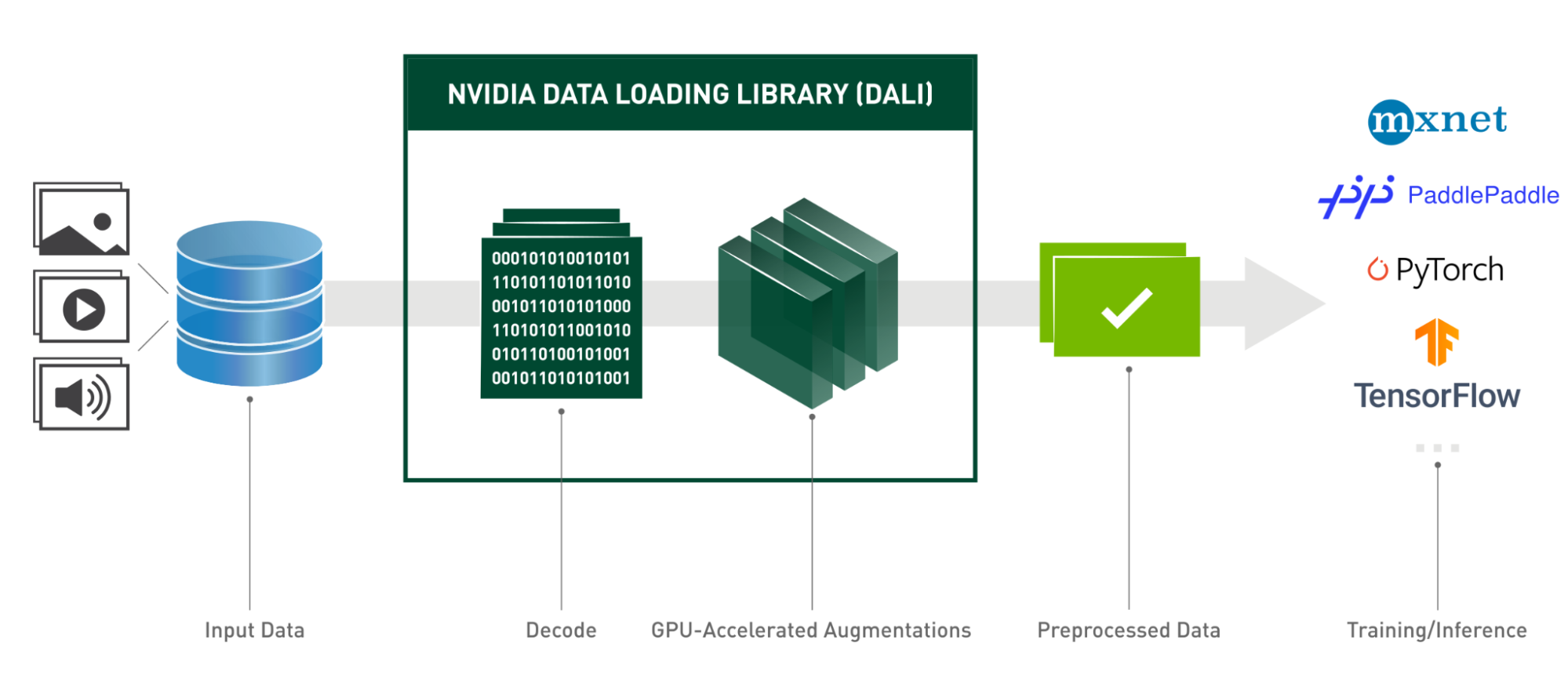
DALI offers an efficient solution for Lightning Pose, by:
- reading the videos.
- handling the decoding process (thanks to the NVIDIA Video Codec SDK).
- applying various augmentations (rotation, resize, brightness, and contrast adjustment, or even adding shot noise).
Using DALI, Lightning Pose increases training throughput for video data and maintains the desired performance of the whole solution by fully using GPUs.
DALI can also be combined with additional data loaders working in parallel. The International Brain Laboratory, a consortium of 16 different neuroscience labs, is currently integrating DALI loaders to predict poses in 30,000 neuroscience experiments.
The benefit of open-source cooperation
The research is a great example of value created by the cooperation of the open-source community. DALI and Lightning Pose, both open-source projects, are highly responsive to community feedback and inquiries on GitHub. The collaboration between these projects began in mid-2021 when Dan Biderman, a community member, started evaluating DALI technology. Dan’s proactive engagement and the DALI team’s swift responses fostered a productive dialogue, which led to its integration into Lightning Pose.
Download and try DALI and Lightning Pose and DALI; you can reach out to contacts for both directly through their GitHub pages.
Read the study, Improved animal estimation through semi-supervised learning, Bayesian ensembling, and cloud-native open-source tools.

 Gathering business insights can be a pain, especially when you’re dealing with countless data points. It’s no secret that GPUs can be a time-saver for…
Gathering business insights can be a pain, especially when you’re dealing with countless data points. It’s no secret that GPUs can be a time-saver for…
 We were stuck. Really stuck. With a hard delivery deadline looming, our team needed to figure out how to process a complex extract-transform-load (ETL) job on…
We were stuck. Really stuck. With a hard delivery deadline looming, our team needed to figure out how to process a complex extract-transform-load (ETL) job on…






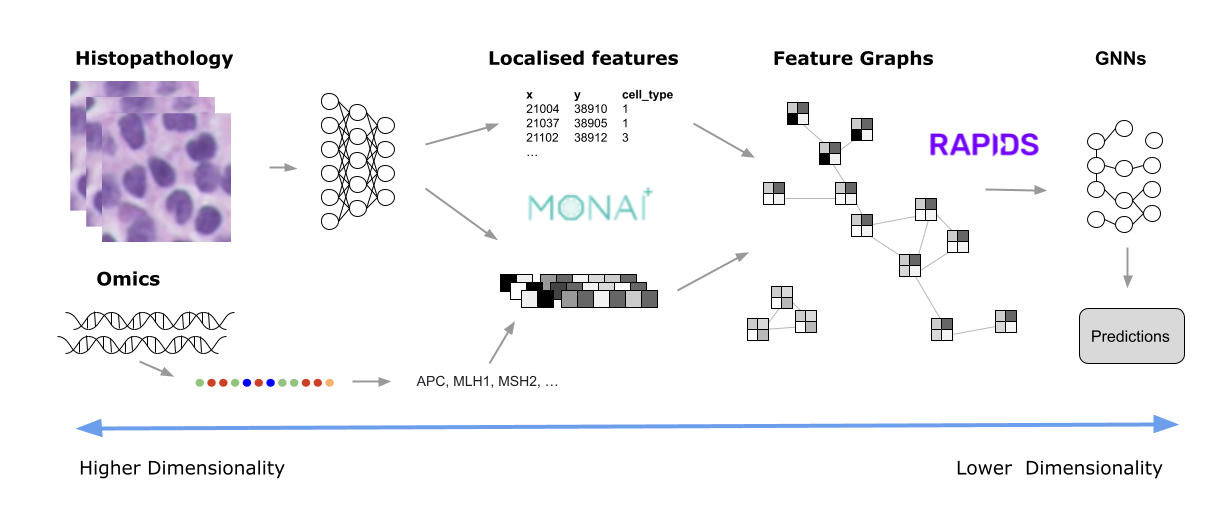
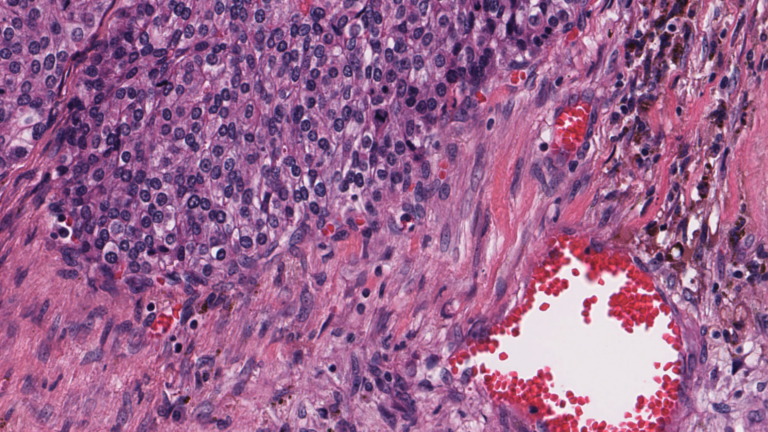 Digital pathology slide scanners generate massive images. Glass slides are routinely scanned at 40x magnification, resulting in gigapixel images. Compression…
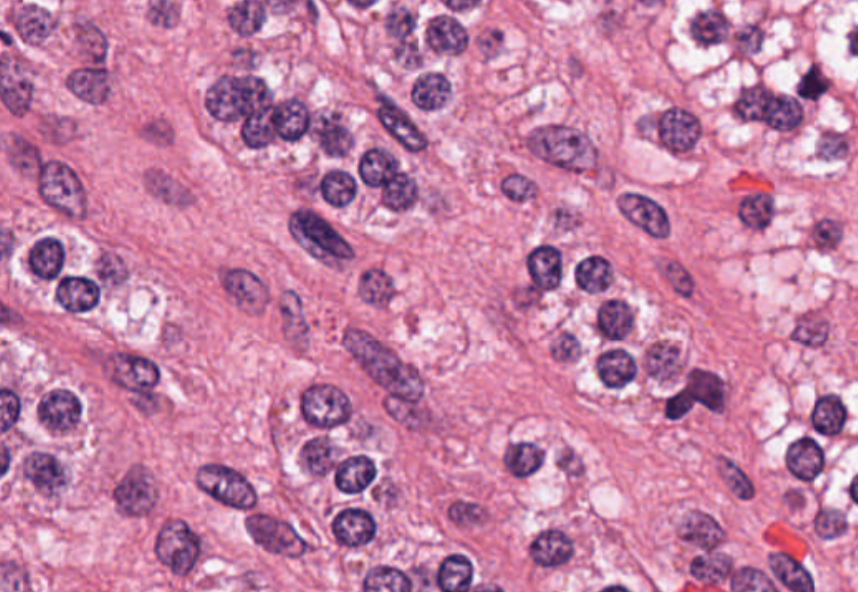
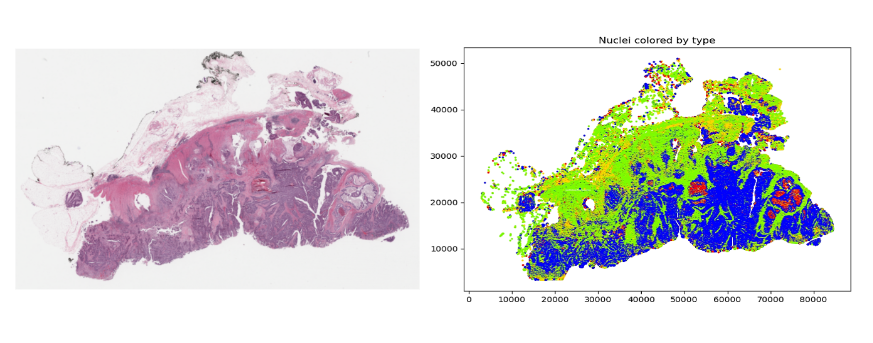
Digital pathology slide scanners generate massive images. Glass slides are routinely scanned at 40x magnification, resulting in gigapixel images. Compression…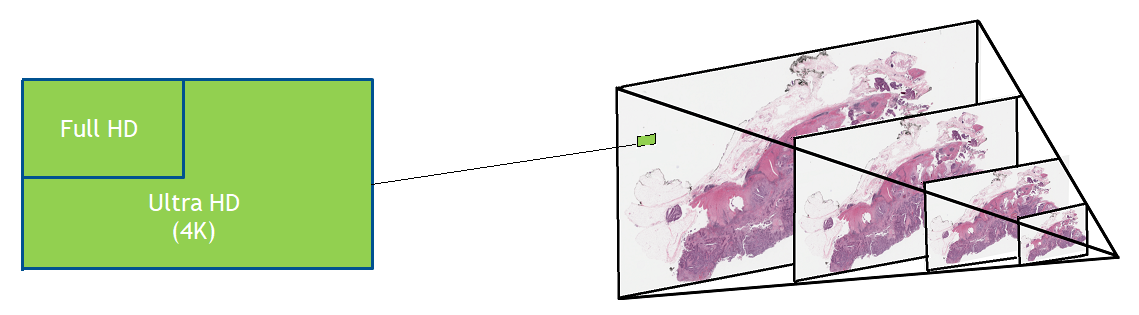
 NVIDIA SDK Manager is the go-to tool for installing the NVIDIA JetPack SDK on NVIDIA Jetson Developer Kits. It provides a guided and simple way to install the…
NVIDIA SDK Manager is the go-to tool for installing the NVIDIA JetPack SDK on NVIDIA Jetson Developer Kits. It provides a guided and simple way to install the…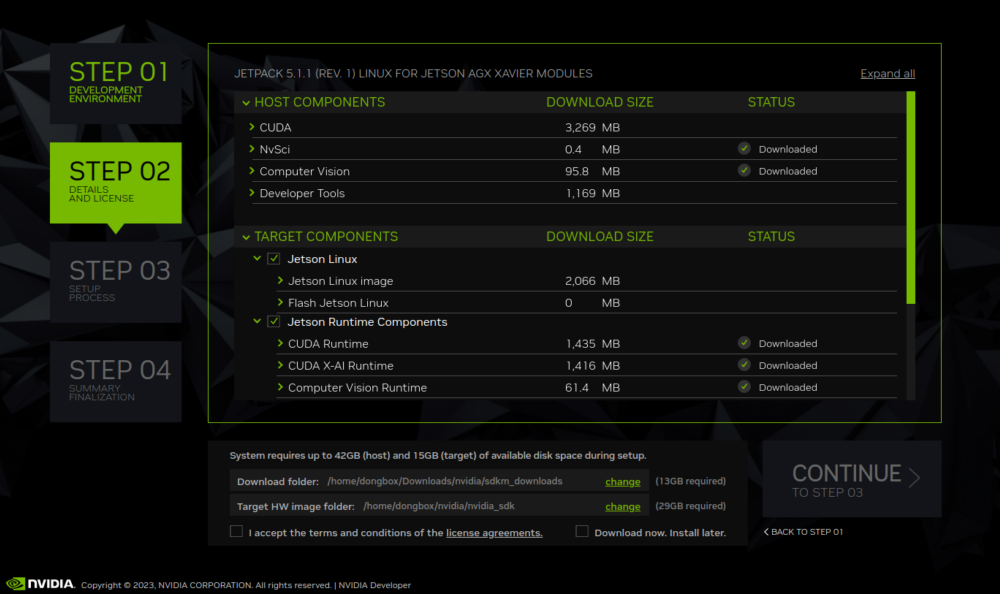
 Gain insights from advanced AI use cases powered by the NVIDIA Jetson Orin in ruggedized environments.
Gain insights from advanced AI use cases powered by the NVIDIA Jetson Orin in ruggedized environments.